In re-examining my preliminary data on Wednesday, I realized I had collected some positive results! I had run a set of samples through a QRT-PCR reaction on Tuesday with the new primers that I received on Monday. These samples were from my experimental assays and represented different time points and test conditions. When I looked at the results on Tuesday, I didn't think much of them, other than the fact that my no template controls (the reactions that are supposed to be blanks) gave me some product, indicating the reactions are still not 100% clean.
But when I went back to the numbers the computer gave me for my test samples, I realized I overlooked the initial data. This is because the data that the program gives me is sort of backwards and not entirely intuitive in the way that I'm using it. The program gives me cycle threshold (Ct) values (marked by the dotted red line in the graph), or the number of cycles that it took a sample to reach a certain threshold in terms of amplified product. This allows us to directly compare the amount of starting product (cDNA) between two samples. For instance, if sample A has a Ct value of 25.88 and B has a Ct of 27.34, sample A had more starting template since it crossed the threshold at an earlier cycle. This is where it gets confusing. In my mindset, the higher value would have more product, but really, it's the other way around. When comparing these values between different samples and time points, it gets even more confusing in determining whether there are changes in starting template.
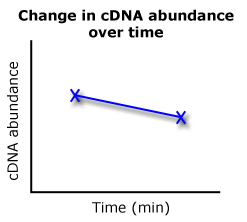
If sample A was the time 0 value and sample B was the time 60 value, we'd essentially get a graph like this on the right. Sample A's Ct of 25.88 is greater in cDNA abundance than sample B's 27.34 Ct. My QPCR comparisons will look a lot like these two cartoons, where I compare Ct values and determine what that data looks like on a graph.
So here I am going back through the data, comparing starting Ct values (0 minutes) with end Ct values (60 minutes) between two different treatments and DNA constructs from my transformed diatoms. Of these eight Ct values (four combinations, two time points each), I realized I had something. My three "controls" all showed the appropriate increase or decrease in mRNA transcript levels, mirroring previously established results by our lab. But when I looked at the experimental line of diatoms, the line I'm most interested, this pattern completely changed from its control. OHHHH YEAHHHH.
 |
| Positive preliminary results: victory! |
 |
| A cartoon representing my results. |
However, I'm not out of the woods yet. I'm still having trouble with my primers and I may need to order the primers a third time. My no template controls are still giving me product, but that may be due to primer dimers. I'm going to run a gel in an attempt to see what is going on, but I may have to eventually sequence the bands to see whether I have some plasmid DNA contamination that I don't know about. All of this will be sorted out, hopefully, sooner or later. But I have some other projects going on that I really should take care of as well. Juggling so many different things is really time consuming, but that's a topic for another time.









